Google-owned artificial intelligence firm DeepMind developed a system AlphaFold to solve the age-old ‘protein folding problem’ or answer the question of how a protein’s amino acid sequence dictates its 3D structure.
Proteins are made up of thousands of amino acids, and millions of small-scale interactions between molecules play into their 3D forms. Understanding the structure of just one protein requires years of work and highly specialized equipment. Thus, researchers have grappled with the complexity of protein folding for decades.
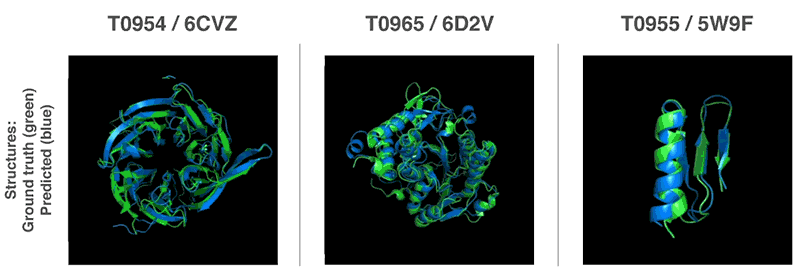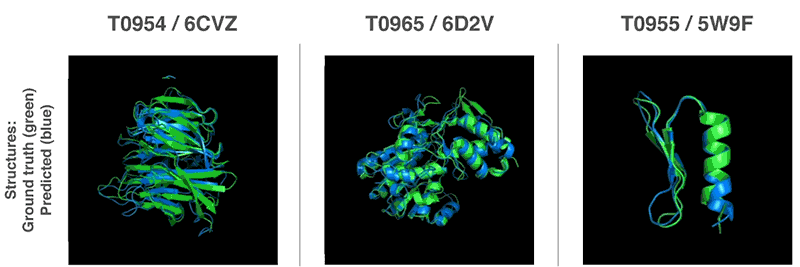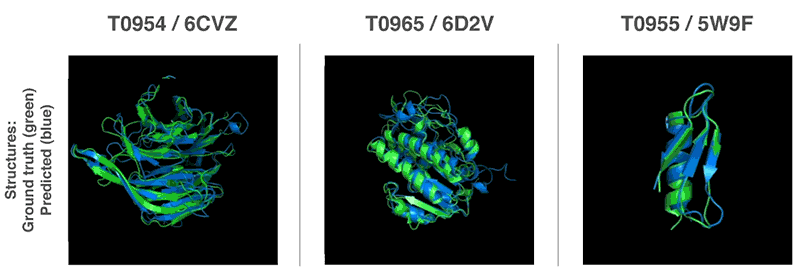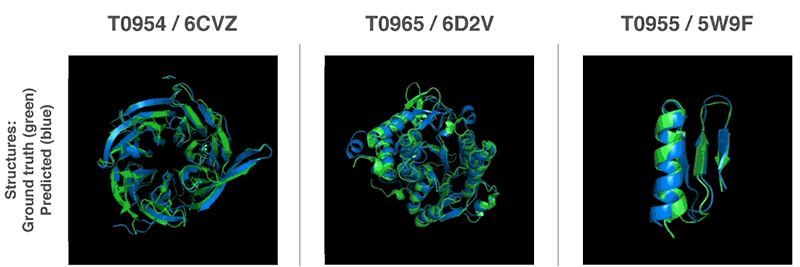
AlphaFold was trained on data from 170,000 known proteins, whose structures were deciphered the traditional way. Now, this technology has an average accuracy score of 92.4 out of 100 for predicting protein structure, and a score of 87 for more complex architectures.
Almost all diseases, such as Alzheimer’s disease, cancer, and COVID-19, involve protein structure, so AI opens the door for faster drug development and a better understanding of the biological processes underlying these health conditions.
Winner of the 2009 Nobel Prize in Chemistry, Venki Ramakrishnan, remarked, “This computational work represents a stunning advance on the protein-folding problem, a 50-year-old grand challenge in biology. It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research.”
It will take some time to improve the algorithm’s predictive power and to bridge the gap between computer modeling and real-world pharmaceutical implementation, but AlphaFold will undoubtedly deepen our understanding of the role protein folding plays in a myriad of diseases.
Outside of medicine, AlphaFold could identify enzymes that break down plastic waste or capture carbon dioxide from the atmosphere, a useful tool in the long-standing battle against climate change.